For lacking words and/or complementary information, please use the glossaries made by Slice et al.:
- http://life.bio.sunysb.edu/morph/glossary/gloss1.html
- http://life.bio.sunysb.edu/morph/glossary/gloss2.html
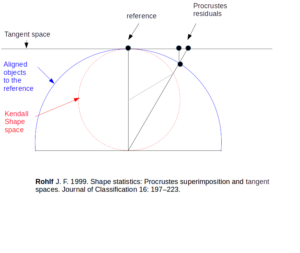
- Aligned coordinates (or specimens, individuals, objects, configurations)
Also called “rotated” objects, or aligned objects are defined by their residual coordinates after GPA, i.e., the coordinates resulting from the Procrustes superimposition to the consensus object (translation, scaling and rotation).
XYOM shows you these objects if you ask for the ROTATED graphic after GPA.
- Hierarchical clustering analysis, single linkage algorithm
Shortly, it is what allows you to build a classification tree, also called a dendrogram. It has no phylogenetic ambition, since it does not use an outgroup to compute the topology. However, assuming the morphometric traits contain a strong phylogenetic signal, the dendrogram information brings a first vision of evolutionary relationships between OTUs.
XYOM uses the Single Linkage Hierarchical Clustering algorithm.
Next version will add the UPGMA algorithm, less sensible to outliers.
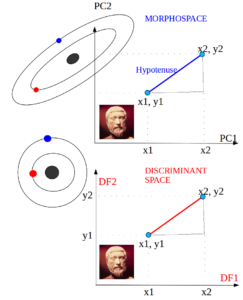
- Mahalanobis distance
The Mahalanobis distances (D2) is the Euclidean distance between group centroids measured in the discriminant space, i.e., on a scale that is adjusted to the (pooled) within-group variance [W] in the direction of the group difference [B]. …
[inverse W] x [B]
The Mahalanobis distance is essentially a distance in a geometric space in which the variables are uncorrelated and equally scaled. …
(Strauss, Discriminating Groups of Organisms. 73-91)
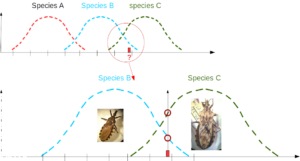
- Maximum likelihood method
What is the chance of observing particular data (or sample) in a given reference group? Comparing this probability among various reference groups allows one to propose a classification, hence to suggest an identification by selecting the reference group having the highest likelihood to fit the data.
In the example below, red interrogation mark represents a morphometric trait which fits the known range of species B and C. Its value seems more frequent in species C.
See also here.
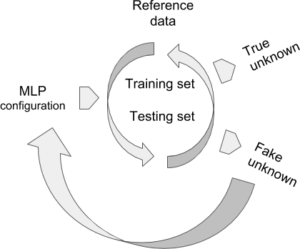
- Multilayer Perceptron (MLP)
MLP configuration: normalising input data or not, number of hidden layers, number of neurons by hidden layer, learning rate, accepted error at training, accepted score at the validation test.
Reference data (raw data by default, or optionally normalized) are documented by the subdivision argument because they are supposed to contain at least two different groups. During the learning step, they are organized iteratively into training and validation (testing) sets. The training step depends on the learning rate and the admitted error. The testing set is validated by XYOM if the score of correct assignment > 75%.
Fake unknown is an optional step, currently imposed by XYOM. The user knows the identity of these “unknown”, which allows for a last validation step on specimens that were not part from the reference data, i.e., untrained data. If the results look unsatisfactory, please try the MLP again modifying its default configuration, for instance normalising the input data, or reducing (or increasing) the number of neurons.
True unknown If validated, the weights produced by the MLP can be used to tentatively identify unknown individuals (see XYOM, IDENTIFICATION).
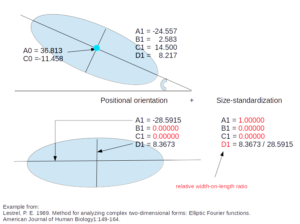
- Normalized Elliptic Fourier coefficients (NEF)
Fourier coefficients (after Elliptic Fourier Analysis) corrected for size and orientation. For each object, size estimator for the scaling is (half) the grand axis of the first ellipse (A1).
- Orthogonal Projections (ORP)
Also called “Tangent space variables”, or “Procrustes residuals”. They are aligned objects orthogonally projected onto the flat plane tangent to the consensus of the aligned objects. These variables are in an Euclidean space, so that they can be used for multivariate statistics like PCA or DA.
- Overfitting
The computation of weights (coefficients to be applied for each input variable) is performed on a limited reference material – even if abundant or huge, it is limited. An excess of accuracy could result in a reduced capacity to generalize the performance to other sets of data.
That is why the testing set is validated by XYOM before to reach very high accuracy, stopping the learning process when the score of correct assignment > 75%.
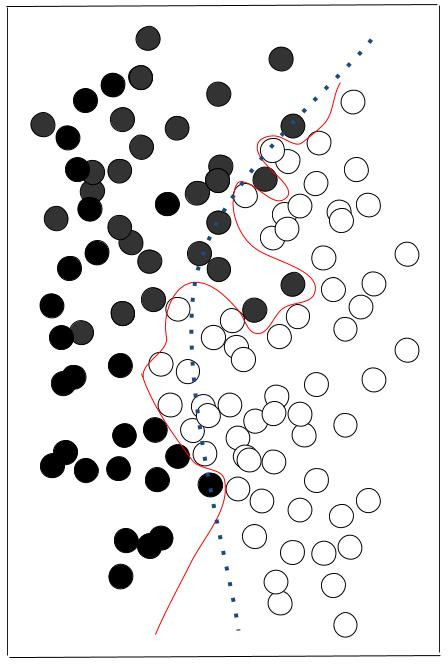
Overfitting illustration. The red line perfectly separates black and white data, but is specialised in these data, and won’t be adapted to other sets of black and white data. The dashed green line is less performant but more likely to adapt to other situations.
- Procrustes residuals, Tangent space variables
See Orthogonal Projections
- Residual Coordinates, Rotated (specimens, individuals, objects, configurations)
See “Aligned” coordinate
- Subdivision
XYOM asking for the “subdivision” is related to the input file arrangement of your data matrix. The data frequntly correspond to different groups: for XYOM (and for CLIC), they must be arranged sequentially: in the input file, each group is followed by another group.
For instance, if the input file is about three species, A, B and C, the individuals of A must be grouped in a submatrix A, followed by the submatrix B, followed by the submatrix C.
Thus, XYOM asks not only for the input file, but also for its subdivision. In the example above, a subdivision like “20,24,29” means that the 20 first rows are about individuals of species A (rows 1 to 20), the next 24 rows about individuals of species B (rows 21 to 44), and the next 29 rows about species C (rows 45 to 73).
If there is no subdivision, please indicate the total number of rows of the input data file.
- Validated classification, Cross Checked Classification (CCC), ‘‘leave one out’’, jackknife:
In this kind of classification process, each individual is iteratively excluded from the analysis to see how re-sampling the dataset affects the assignment estimation.
XYOM provides various methods for validated classification (called CCC – for cross checked classification – in the CLIC package)
- The Maximum likelihood method CCCMli
- The Mahalanobis distance method CCCMaha
- The between group PCA method (for next XYOM version) CCCbgPCA
- The machine learning (supervised) method with the Multiplayer Perceptron (MLP)